
Mosquitos + Tensors + Genetics + CS + Networks + Math + Coffee
Developed in John Marshall's Lab by:
Lead: Héctor M. Sánchez C.
Core Dev: Sean L. Wu, Jared Bennett
Ecology: Tomás León
Spatial Analysis: Chris De León
Data Analysis: Priscilla Zhang
Optimization: Valeri Vasquez
Former Members: Gillian Chu, Maya Shen, Yunwen Ji, Víctor Ferman, Biyonka Liang, Sarafina Smith, Sabrina Wong, Chase Violet
Mosquito Gene Drive Explorer
Brief Description
MGDrivE is a framework designed to serve as a testbed in which gene-drive releases for mosquito-borne diseases control can be tested. It is being developed to accommodate various mosquito-specific gene drive systems within a population dynamics model that allows migration of individuals between nodes in a spatial landscape.
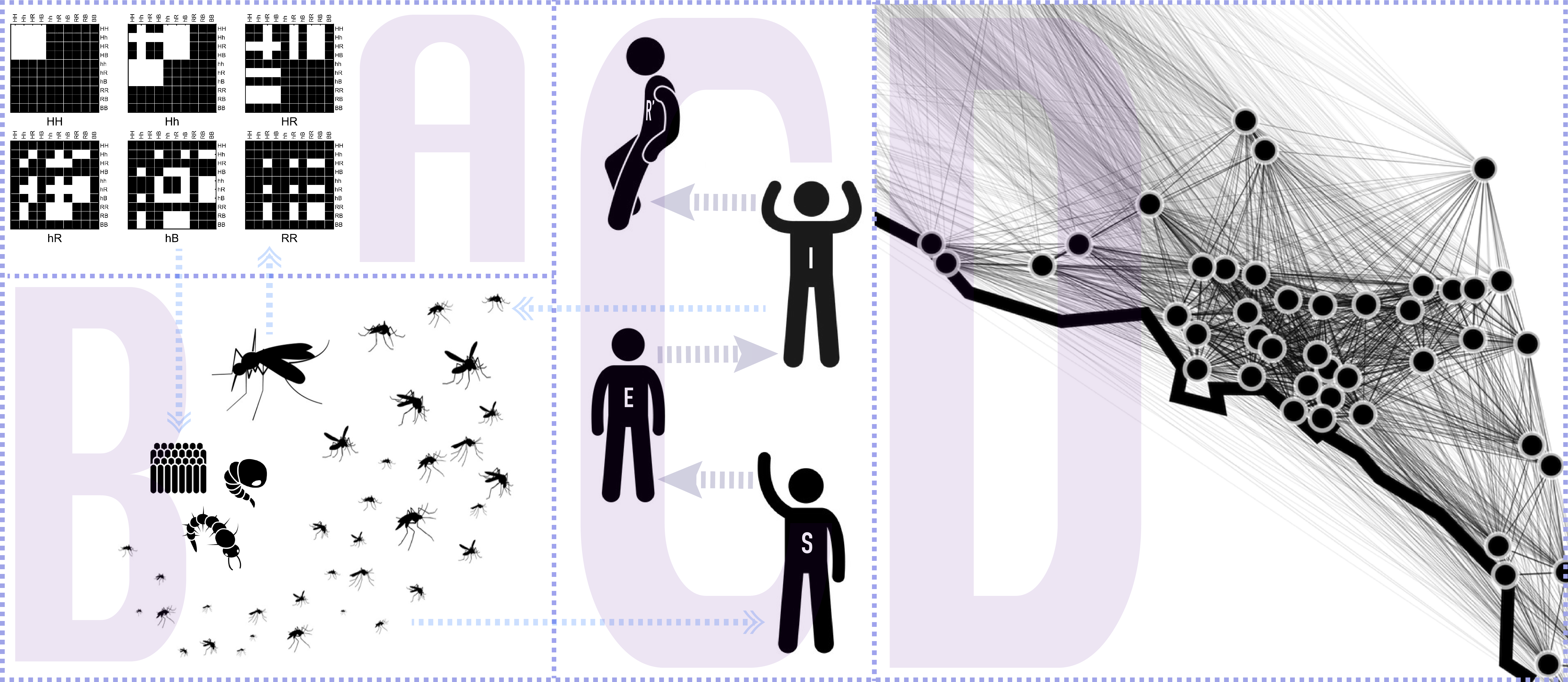
Currently, the model contains three modules: inheritance (A), life-history (B), and migration (D); while an epidemiological module (C) is currently under development for our upcoming v2.
Demonstration
In this demo, we are releasing a total of 100 mosquitoes homozygous for the CRISPR/CAS9 and one with a mutation that makes the mosquito resistant to the construct. Each node in the network represents a mosquito population laid down in a spatial scenario (this could be though of as a household, house block or even city if needed). We simulate how the genetic construct would propagate across the nodes of the network if mosquitoes were slowly migrating between populations with a probability based on proximity. To watch more videos take look at our youtube playlist.
How does it work?
The main idea behind this model is to consider the inheritance matrix of genotypes a three-dimensional structure in which each intersection point determines the ratio/probability of a specific offspring genotype (z axis) provided that a certain combination of male-female genotypes (x and y axis). This allows us to use tensors as the basis for our calculations which has many advantages, some of them being: computational speed, model's transparency and extendability.

The second novel idea in our framework is to consider the spatial layout as a network of inter-connected breeding habitats (D). By performing this abstraction we are able to transform these landscapes into distances matrices, and then into transition probabilities matrices (through the use of movement kernels). This allows our framework to be able to model arbitrary topologies in which we can simulate mosquito populations mating and migrating in realistic geographical settings.

Installation
Our package is now available on the CRAN repository so it can be easily installed and imported with the following commands:
install.packages("MGDrivE")
library(MGDrivE)
To facilitate the data analysis of the results produced by MGDrivE, we provide a python package MoNeT_MGDrivE installable through pip:
pip install MoNeT-MGDrivE
This package provides all the functions we have used to perform the analyses on spatio-temporal gene-drives spread, and is part of our companion project: MoNeT, so have a look at the project's website for information on some of the work we are doing!

Publications and Presentations
- [Preprint]: Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. biorxiv. Adriana Adolfi, Valentino M. Gantz, Nijole Jasinskiene, Hsu-Feng Lee, Kristy Hwanga, Emily A. Bulger, Arunachalam Ramaiahd, Jared B. Bennett, Gerard Terradas, J.J. Emerson, John M. Marshall, Ethan Bier, Anthony A. James (2020).
- [Preprint]: A home and rescue gene drive forces its inheritance stably persisting in populations. biorxiv. Nikolay P. Kandul1, Junru Liu1, Jared B. Bennett3, John M. Marshall2, and Omar S. Akbari (2020).
- [Preprint]: Inherently confinable split-drive systems in Drosophila. biorxiv. Gerard Terradas, Anna B.Buchman, Jared B.Bennett, Isaiah Shriner, John M.Marshall, Omar S.Akbari, Ethan Bier (2020).
- [Preprint]: MGDrivE 2: A simulation framework for gene drive systems incorporating seasonality and epidemiological dynamics. biorxiv. Sean L. Wu, Jared B. Bennett, Héctor M. Sánchez, Andrew J. Dolgert, Tomás M. León, John M. Marshall (2020).
- [Paper]: Molecular Cell. Active Genetic Neutralizing Elements for Halting or Deleting Gene Drives. Molecular Cell. Xiang-Ru Shannon Xu, Emily A. Bulger, Valentino M. Gantz, Carissa Klanseck, Stephanie R. Heimler, Ankush Auradkar, Jared B. Bennett, Lauren Ashley Miller, Sarah Leahy, Sara Sanz Juste, Anna Buchman, Omar S. Akbari, John M. Marshall, Ethan Bier (2020)
- [Paper]: Modeling confinement and reversibility of threshold-dependent gene drive systems in spatially-explicit Aedes aegypti populations. BMC Biology. Héctor M. Sánchez C., Jared B. Bennett, Sean L. Wu, Gordana Rašić, Omar S. Akbari, John M. Marshall (2020).
- [Paper]: A transcomplementing gene drive provides a flexible platform for laboratory investigation and potential field deployment. Nature Communications. Víctor López Del Amo, Alena L. Bishop, Héctor M. Sánchez C., Jared B. Bennett, Xuechun Feng, John M. Marshall, Ethan Bier, Valentino M. Gantz (2020).
- [Paper]: Development of a confinable gene drive system in the human disease vector Aedes aegypti. eLife. Ming Li, Ting Yang, Nikolay P Kandul, Michelle Bui, Stephanie Gamez, Robyn Raban, Jared Bennett, Héctor M Sánchez C, Gregory C Lanzaro, Hanno Schmidt, Yoosook Lee, John M Marshall, Omar S Akbari (2020).
- [Paper]: MGDrivE: A modular simulation framework for the spread of gene drives through spatially-explicit mosquito populations. Methods in Ecology and Evolution. Hector M. Sanchez C., Sean L. Wu, Jared Bennett, and John M. Marshall (2019).
- [Paper]: Transforming Insect Population Control with Precision Guided Sterile Males. Nature Communications. Nikolay Kandul, Junru Liu, Héctor M. Sánchez C., Sean L. Wu, John M. Marshall, Omar S. Akbari (2018).
- [Paper]: Consequences of Resistance Evolution in a Cas9-Based Sex-Conversion-Suppression Gene Drive for Insect Pest Management. Proceedings of the National Academy of Sciences of the United States of America. Karaminejadranjbar, Mohammad, Kolja N Eckermann, Hassan M M Ahmed, and Héctor M Sánchez C, Stefan Dippel, John M. Marshall, Ernst A. Wimmer (2018)
- [Paper]: Transforming Insect Population Control with Precision Guided Sterile Males. Nature Communications. Nikolay Kandul, Junru Liu, Héctor M. Sánchez C., Sean L. Wu, John M. Marshall and Omar S. Akbari (2018).
- [Paper]: Overcoming evolved resistance to population-suppressing homing-based gene drives. Nature Scientific Reports. John M. Marshall, Anna Buchman, Héctor M. Sánchez C., Omar S. Akbari (2017)
- [Thesis]: Integrating Synthetic Biology Derived Variables into Ecological Risk Assessment Using the Bayesian Network – Relative Risk Model: Gene Drives to Control Nonindigenous M. musculus on Southeast Farallon Island. Ethan A. Brown (2020).
- [Thesis]: Quantifying the Relationship Between Spatial Habitat Distribution and Homing Allele Fixation in MCR Gene Drive Systems for Aedes aegypti Mosquitoes. Biyonka Liang, Héctor M. Sánchez C., Sean L. Wu, John M. Marshall, Christopher Pacioreck (2019).
- [Talk]: MGDrive: Landscape Clustering. NCUR. Gillian Chu, Héctor M. Sánchez C (2020).
- [Talk]: Space, The Final Frontier: Mosquito Movement and Vector Control (MoNeT + MGDrivE). MASH-MAP Malaria Retreat. Héctor M. Sánchez C., John M. Marshall, David L. Smith (2019).
- [Talk]: MGDrivE: The Original Trilogy. Computational Biology Retreat. Héctor M. Sánchez C., Jared B. Bennett, Valeri Vasquez, Sean L. Wu, Tomás León, Sarafina Smith, Gillian Chu, John M. Marshall (2018).
- [Talk]: MGDrivE: A simulation framework for gene drive releases in spatially explicit mosquito populations, and its application to mosquito borne diseases control. Infectious Diseases Dynamics Conference. Héctor M. Sánchez C., Jared B. Bennett, Sean L. Wu, Valeri Vasquez, John M. Marshall (2018).
- [Talk]: MGDrivE: A simulation framework for gene drive in spatially explicit mosquito populations and its application to threshold-dependent systems. Annual Meeting of the Society for Mathematical Biology and the Japanese Society for Mathematical Biology. Héctor M. Sánchez C., Sean L. Wu, Jared B. Bennett, John M. Marshall (2018).
- [Talk]: MGDrivE: A Mosquito Population Framework to Evaluate and Optimize Gene-Drive Releases for Vector-Borne Diseases Control. NorCal CompBio Symposium. Héctor M. Sánchez C., Sean L. Wu, Jared Bennett, John M. Marshall, (2017).
- [Lightning Talk]: MGDrivE: Its application to releases optimization, and confinement in an Australian setting. NorCalCompBio Symposium. Héctor M. Sánchez C., Jared B. Bennett, Sean L. Wu, Valeri Vasquez, John M. Marshall (2018).
- [Poster]: MGDrivE: A simulation framework for gene drive in spatially-explicit mosquito populations and its application to threshold-dependent systems. American Society of Tropical Medicine and Hygiene. Héctor M. Sánchez C., Jared B. Bennett, Sean L. Wu, Valeri Vasquez, John M. Marshall (2018).
- [Poster]: A Tale of Two Cities: Confinability and Remediation Potential of UDMel and Translocation Gene-Drives in Spatially Explicit Aedes aegypti Populations. University of California, Berkeley Compitational Biology Retreat. Héctor M. Sánchez C., Sean L. Wu, Jared Bennett, John M. Marshall (2017).










